The Dog Days of PySpark

Photo by Mohammad Mardani on Unsplash
PySpark. One of those things to hate and love, well … kinda hard not to love. PySpark is the abstraction that lets a bazillion Data Engineers forget about that blight Scala and cuddle their wonderfully soft and ever-kind Python code, while choking down gobs of data like some Harkonnen glutton.
But, that comes with a price. The price of our own laziness and that idea that all that glitters is gold, to take the easy path. One of the main problems is the dreadful mistake of mixing native Python in with your PySpark and expecting things to go fine at scale. Which it most assuredly will not.
PySpark example with native Python.
So what am I exactly talking about? Well, I would be more than happy to give you a tirade on the curses of UDFs and how 99% of them are unneeded, and wreaking havoc on pipelines around the world, instead, I want to give you a simple example. Maybe something you’ve never thought of, maybe so, we will see.
Row-wise computation in PySpark.
Let’s start with a simple example. Let’s say we have a Dataframe with 1.5 billion records, with 8 integer columns.
from pyspark.sql import SparkSession
from pyspark.sql.functions import rand
# Initialize Spark Session
spark = SparkSession.builder \
.appName("Generate Large DataFrame") \
.getOrCreate()
# Set the number of rows and partitions
num_rows = 1_500_000_000
num_partitions = 5 # Adjust this value based on the resources available
# Define the range for random integer values
min_value = 0
max_value = 100
# Create a DataFrame with random integer values for each column
df = spark.range(0, num_rows, numPartitions=num_partitions)
# Generate random integer values for each column using the 'rand()' function
df = df.select(
(rand() * (max_value - min_value) + min_value).cast("integer").alias("col1"),
(rand() * (max_value - min_value) + min_value).cast("integer").alias("col2"),
(rand() * (max_value - min_value) + min_value).cast("integer").alias("col3"),
(rand() * (max_value - min_value) + min_value).cast("integer").alias("col4"),
(rand() * (max_value - min_value) + min_value).cast("integer").alias("col5"),
(rand() * (max_value - min_value) + min_value).cast("integer").alias("col6"),
(rand() * (max_value - min_value) + min_value).cast("integer").alias("col7"),
(rand() * (max_value - min_value) + min_value).cast("integer").alias("col8"),
)
# Show the first 10 rows of the DataFrame
df.show(10)
So in real life, we would probably have a few more columns, maybe a bunch of random STRING columns as part of this Dataframe. Also, let’s just say you had a requirement to add an additional column that was the sum of these other integer columns by row. No grouping.
Let’s say there are 15 STRING columns and you don’t want to GROUP BY each one of those 15 STRING columns to simply get an additional column added. Also, to throw an extra curve ball into the mix, the Dataframe can change and so the columns you would like to add are variable. Aka, sometimes it’s a list of 5 columns, sometimes is a list of 8, who knows?
So, you in all your genius decide to Google for the answer, say “row-wise addition operation in PySpark over multiple columns.”
You will get multiple results, some of them from our beloved Stackoverflow, like this number one result in Google. Basically, if you are not careful and in a hurry, you will be encouraged to use Python’s built-in summethod, and maybe reduce.
The Native Python way.
So let’s import the new tools we need.
from functools import reduce
from operator import add
from datetime import datetime
from pyspark.sql.functions import rand, col
And we can add this simple line to our code.
df = df.withColumn(“result” ,reduce(add, [col(x) for x in df.columns]))
I mean, it’s straightforward, and easy, right? Makes sense. Of course, we could qualify our list compression with our own list of columns, so we only do what we want. I of course timed the run. We write out the results to ensure our 500 million records all get the computation.
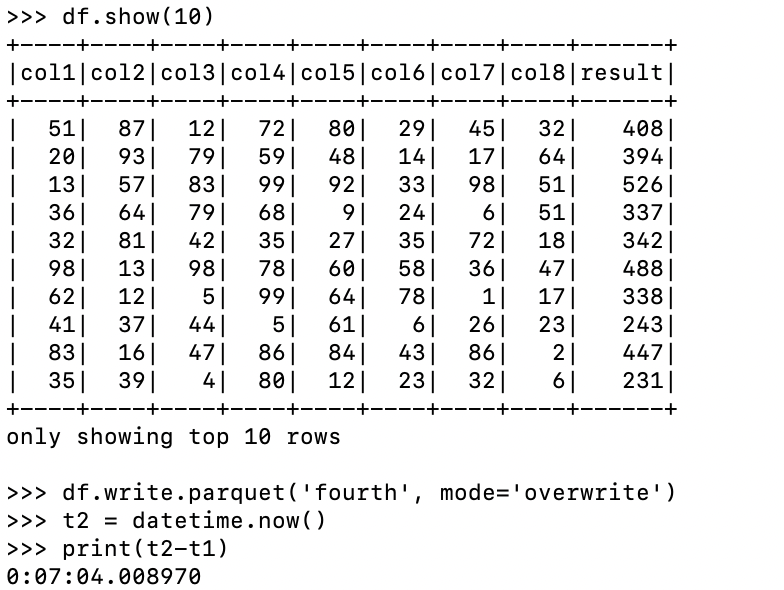
So, a little over 7 minutes! I’m no Spark expert, but I expect using native Python as such is going to require some data serialization and de-serialization between the Python processes and Spark JVM. But, is it enough extra work to be materially slower than using an expr or selectExpr to solve the problem?
The Spark way.
So, at all costs, we should use Spark to solve the computation, that’s what the experts tell us. Let’s use a expr to solve this problem.
We can simply get a list of the columns we want to add, joining them all together as strings with a + between.
addy = ‘+’.join([str(c) for c in df.columns])
Then we can pass the String into the PySpark function called expr.
df = df.withColumn(“result” , expr(addy))
Also, let’s write out to a different location so this code doesn’t have to overwrite the other dataset.
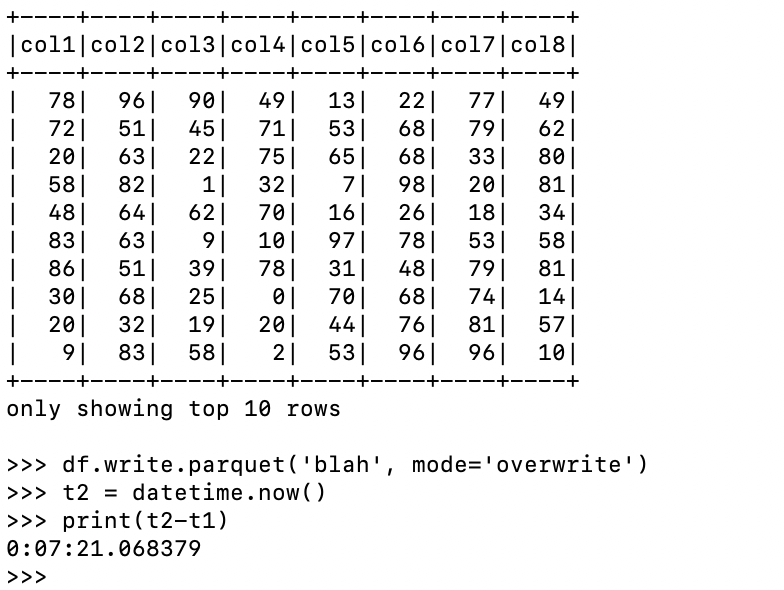
Slower? Well, I didn’t expect that one. We’ve been told a million times over that mixing normal Python in with your PySpark script is the devil’s work. Sure my random number math could have been larger for the second time, but it still doesn’t explain why doing the work with an expr is slower than using add and reduce via Python. Strange.
Is it because of some other Spark setting, partitions perhaps, or the memory configurations? I suppose I could spend more time messing and figuring it out … but … I don’t really feel like it.
Conclusion.
I don’t know.

Hey man, I think you are essentially doing the same thing in both of your examples. In the first one, you are generating the “expressions” using the Python API whereas in the second example, you are generating an SQL string. But both have the same effect. There is no serialization or deserialization, the “col(a) + col(b) … + col(n)” or “a + b + … n” are just converted to some common representation and executed on the workers (on the JVM, Python is not needed AFAIK).
Using Python would be discouraged in case of using an UDF like this:
@udf(returnType=IntegerType()
def sum(some_arg):
# do some summing, etc.
return 10
and then using this UDF like this:
df.withColumn(“col”, sum(col(“input_col”)))
When using a Python UDF, Python interpreter has to be running on the Spark workers. First some data-munching will happen using Scala workers, then the workers will serialize the “input_col” data and send it to the Python interpreter. The interpreter will deserialize the data, perform the UDF, serialize the data and send it back to the Scala worker. And the Scala worker will return the result. So that’s where the overhead would be coming from.
Of course, I think with Apache Arrow this whole serialization and deserialization process might not be needed (but I don’t know much about that). But I guess you would still incur some overhead because you would be executing Python which is slower that Scala.
But using DataFrame API, as far as I know, you are not even using the Python interpreter on the Spark workers.
Also, I think the difference in the results might be just due to chance 🙂
Everything I said might not be 100 % correct but I think generally it should be like this 😀 Love your blog btw, especially the info on Databricks!
Without diving into config/partitions/resources, I’m curious on your benchmarking methodology, was this just 1:1 run comparison or average?
To me, `df.withColumn(“result”, sum([col(c) for c in df.columns]))` is actually the most PySpark-y way.