String Slicing Performance – Python vs Scala vs Spark.

Good ole’ string slicing. That’s one thing that never changes in Data Engineering, working with strings. You would think we would all get to row up some day and do the complicated stuff, but apparently you can’t outrun your past. I blame this mostly on the data and old schools companies. Plain text and flat files are still incredibly popular and common for storing and exporting data between systems. Hence string work comes upon us all like some terrible overload. The one you should fear the most is fixed width delimited files. I ran into a problem recently where PySpark was surprisingly terrible at processing fixed with delimited files and “string slicing.” It got me wondering … is it me or you?
Fixed width delimited text files.
All code available on GitHub.
Normally most plain text and flat files that data is exported in might be csv with commas delimited records, I mean of course their is the ever present and strange tab delimited files with the ole \t. I mean even the pipe delimiter, |, between column values is better then nothing ….
But fixed width?? Seriously?
All this means is that between records in the file, there is a fixed number of spaces, that can be different between records. For example ….
header_1 header_2 header_3 header_4
123 456 789 101112So in the above example the first column starts at position 0 and goes until 8, the second column starts to 9 and goes until 26 …. and so on. Fixed width. Behold the most terrible and evil flat file ever devised by mankind.
String Slicing and Peformance
The problem with files like this is that string slicing, which is pretty basic, becomes one of the only obvious ways of breaking down these delimited files into individual data points to be worked on. And when you have 100+ columns and many millions of rows, this becomes an intensive process.
Why? Because you have to read every row and use your poor CPU to figure out each data point. Also, since you could have hundreds of columns and millions of records in a single file … how do you apply distributed computing to such a problem, since only one machine can read one file at a time? This is a problem.
First, let’s just compare Python vs Scala vs Spark in this simple string slicing operation, see who can process file(s) the fastest, and then try to think outside the box on how we might solve this problem more creatively.
Converting CSV files into Fixed Width nightmares.
We are going to use the Divvy Bike trip data set, I downloaded 21 CSV files and pretend like they are fixed with delimited files. We also going to pretend like they are all a single file for arguments sake.
The files look like this ….
delimited_files = [
'data/202004-divvy-tripdata.csv',
'data/202005-divvy-tripdata.csv',
'data/202006-divvy-tripdata.csv',
'data/202007-divvy-tripdata.csv',
'data/202008-divvy-tripdata.csv',
'data/202009-divvy-tripdata.csv',
'data/202010-divvy-tripdata.csv',
'data/202011-divvy-tripdata.csv',
'data/202012-divvy-tripdata.csv',
'data/202101-divvy-tripdata.csv',
'data/202102-divvy-tripdata.csv',
'data/202103-divvy-tripdata.csv'
]
def convert_csv_to_fixed_width(files: list):
with open('fixed_width_data.txt', 'w') as f:
for file in files:
rows = read_with_csv(file)
for row in rows:
num_rows = range(13)
str_line = ''
for num in num_rows:
value = row[num]
pad_value = value+' '
str_line += pad_value[:45]
f.write(str_line+'\n')
def main():
convert_csv_to_fixed_width(delimited_files)
if __name__ == '__main__':
main()
What I’m doing above is reading in the all the csv files, creating each record with 50 spaces tacked onto it, then slicing it back down to 45 characters and writing out the line.
You can see below the file turns out in a fixed width format. We now have a single file with 3,489,760 lines and 13 columns. Each record is is 45 characters long, for 13 columns.
Python string slicing performance.
Now that we have our fixed width file. Let’s process in Python and see how long it takes to convert this fixed width file back to CSV.
def read_with_csv(file_loc: str):
with open(file_loc) as csv_file:
csv_reader = csv.reader(csv_file, delimiter=',')
for row in csv_reader:
yield row
def read_file_return_records(file_loc: str) -> iter:
with open(file_loc) as fixed_width_file:
lines = fixed_width_file.readlines()
return lines
def slice_line(line: str):
c1 = line[:45].strip()
c2 = line[45:90].strip()
c3 = line[90:135].strip()
c4 = line[135:180].strip()
c5 = line[180:225].strip()
c6 = line[225:270].strip()
c7 = line[270:315].strip()
c8 = line[315:360].strip()
c9 = line[360:405].strip()
c10 = line[405:450].strip()
c11 = line[450:495].strip()
c12 = line[495:540].strip()
c13 = line[540:585].strip()
return c1, c2, c3, c4, c5, c6, c7, c8, c9, c10, c11, c12, c13
def main():
t1 = datetime.now()
with open('result.csv', 'w') as f2:
with open('fixed_width_data.txt', 'r') as f:
lines = f.readlines()
for line in lines:
c1, c2, c3, c4, c5, c6, c7, c8, c9, c10, c11, c12, c13 = slice_line(line)
f2.write(f"{c1}, {c2}, {c3}, {c4}, {c5}, {c6}, {c7}, {c8}, {c9}, {c10}, {c11}, {c12}, {c13} \n")
t2 = datetime.now()
x = t2-t1
print(f"it took {x} to process file")You can see the output file is pretty much like our original files, we converted the fixed with values back into a more normal csv file again.
There is nothing particularly special about the Python code, nice and simple. Just read the file in a a bunch of lines of text, then using the known spacing, or fixed width of each column, string slice out the value and trim down the white space.
Below is a sample of the results.csv we wrote the records back out too.
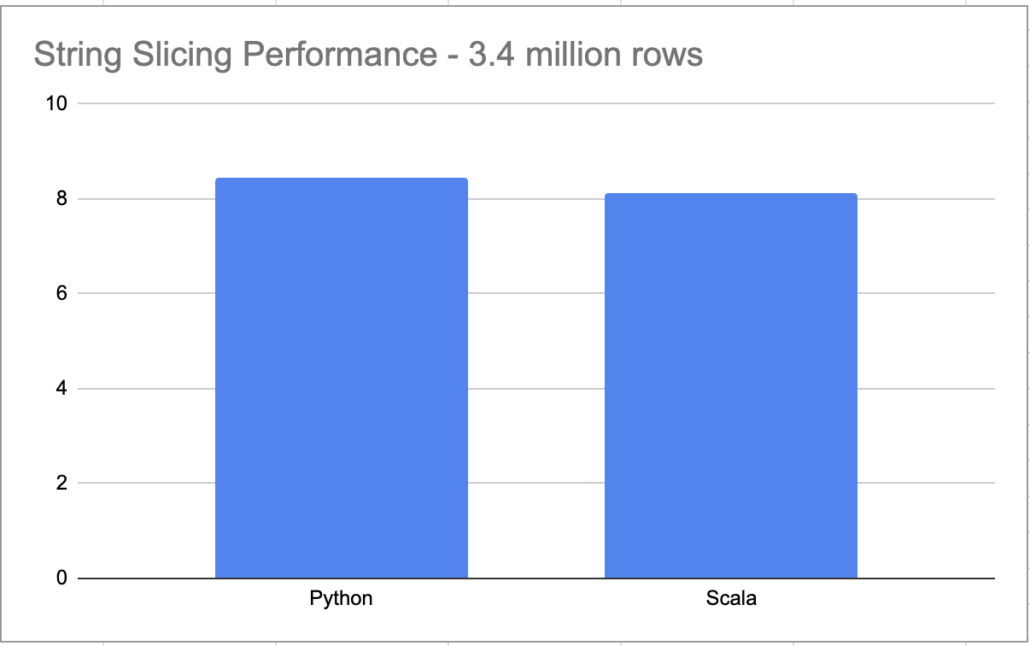
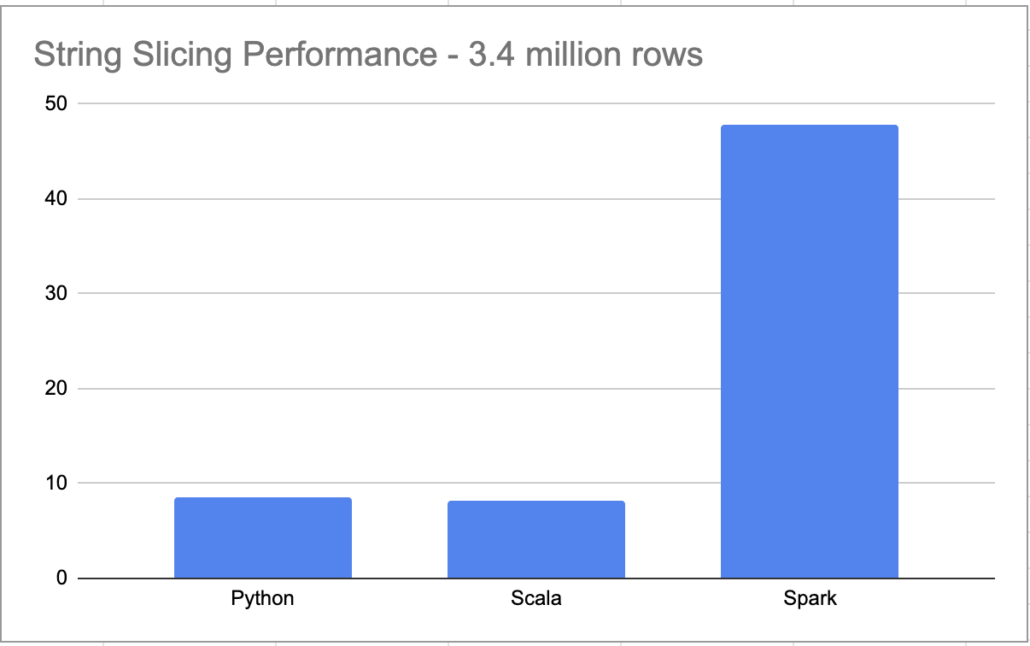
it took 0:00:08.461747 to process file
I know 8 seconds doesn’t seem like a long time, but what if instead of 3.4 million records we had 20 million, and instead of 13 columns we had 113, plus multiple files. You can see where I’m going.
Scala string slicing performance.
I’m terrible at Scala but I am curious if this would be significantly faster re-written with Scala. I will do my best, which isn’t saying much.
I pretty much want to follow the same logic I did with Python, read in a plain text file, bring in all the lines, then slice the column values out of that single string and write it all back out to a file.
import scala.io.Source
import java.io._
object stringSlice {
def main(args: Array[String]): Unit = {
val t1 = System.nanoTime
val fixed_file = Source.fromFile("fixed_width_data.txt")
val result_sink = new File("results.txt")
val bw = new BufferedWriter(new FileWriter(result_sink))
for (line <- fixed_file.getLines){
val (c1, c2, c3, c4, c5, c6, c7, c8, c9, c10, c11, c12, c13) = slice_values(line)
bw.write(s"$c1, $c2, $c3, $c4, $c5, $c6, $c7, $c8, $c9, $c10, $c11, $c12, $c13 \n")
}
bw.close()
val duration = (System.nanoTime - t1) / 1e9d
println(duration)
}
def slice_values(line: String): (String,String,String,String,String,String,String,String,String,String,String,String,String) = {
val c1 = line.slice(0, 45).trim()
val c2 = line.slice(45, 90).trim()
val c3 = line.slice(90, 135).trim()
val c4 = line.slice(135, 180).trim()
val c5 = line.slice(180, 225).trim()
val c6 = line.slice(225, 270).trim()
val c7 = line.slice(270, 315).trim()
val c8 = line.slice(315, 360).trim()
val c9 = line.slice(360, 405).trim()
val c10 = line.slice(405, 450).trim()
val c11 = line.slice(450, 495).trim()
val c12 = line.slice(495, 540).trim()
val c13 = line.slice(540, 585).trim()
(c1, c2, c3, c4, c5, c6, c7, c8, c9, c10, c11, c12, c13)
}
}Not very pretty Scala but it works. I get a good file back out.
and the time …. 8.134520417 to run. Only slightly faster than the Python.
Although it’s probably not fair to add PySpark to the mix, it’s meant for distributed data processing …. the use case I’m thinking about is Spark ETL and Pipelines where you are defaulting to a single technology and getting one or two new files that need to be converted per day.
PySpark string slicing performance.
I’m very curious to see where PySpark will come in to play on the performance side. Of course Spark would win if you had a massive dataset with many thousands or more of files to processes, that’s really what Spark is for. But, like I said, in the real world sometimes you have pipelines that are running all in Spark, and you want to keep it that way for simplicity of the tech stack.
Sometimes you only want to introduce new tools when needed. Let’s see how PySpark handles this single file.
from pyspark.sql import SparkSession
from datetime import datetime
spark = SparkSession.builder.appName('SparkSlice').getOrCreate()
t1 = datetime.now()
fixed_file = '/Users/danielbeach/Desktop/fixed_width_data.txt'
df = spark.read.csv(fixed_file)
df.createOrReplaceTempView('trips')
result = spark.sql("""
SELECT
trim(substring(`_c0`, 0, 45)) as ride_id,
trim(substring(`_c0`, 45, 90)) as rideable_type,
trim(substring(`_c0`, 90, 135)) as started_at,
trim(substring(`_c0`, 135, 180)) as ended_at,
trim(substring(`_c0`, 180, 225)) as start_station_name,
trim(substring(`_c0`, 225, 270)) as start_station_id,
trim(substring(`_c0`, 270, 315)) as end_station_name,
trim(substring(`_c0`, 315, 360)) as end_station_id,
trim(substring(`_c0`, 360, 405)) as start_lat,
trim(substring(`_c0`, 405, 450)) as start_lng,
trim(substring(`_c0`, 450, 495)) as end_lat,
trim(substring(`_c0`, 495, 540)) as end_lng,
trim(substring(`_c0`, 540, 585)) as member_casual
FROM trips;
""")
result.write.mode('overwrite').option("header", "true").csv('results.txt')
t2 = datetime.now()
x = t2-t1
print(f"it took {x}")it took 0:00:47.797910
Yikes. Now I’m sure if we were processing 200 GB+ worth of data, the story might be different. But this just goes to show that when designing pipelines, you have to pick the right tool for the right job. Picking distributed Spark to process and string splice a few files when time matters (because you are paying for it), probably isn’t the best idea.
Musings
I’m a little surprised my crappy Scala code isn’t that much faster then the Python code. You known people complain about how slow Python is. If I’m not really gaining any significant speed up’s using Scala to slice strings, then I will probably just stick with Python.
I know the problem with using PySpark is that there is a lot going on their, the distributed system was made to go through optimizers etc, Spark isn’t that useful when working with a single file. It is a bit annoying when you want to keep your codebase in a single technology stack and not have to swap things out.
Unfortunately running Spark on cluster’s is never really that cheap, so I guess that means for future Spark pipelines where you start to wander off into other things, like single file processing … which is easy to do … you might think twice.
All code seen here is on GitHub.

lots of industries out there with old mainframe processing machines so this kind of data files will still be around to create jobs for Data Engineers not afraid to roll up their sleeves. I doubt black box solutions for DE work could ever successfully process this kind of data. Savvy of you to use Scala for comparison as I’ve always figured Scala was used for low level byte processing work like streaming data parsing.