Performance Testing Postgres Inserts with Python
Sometimes I get to feeling nostalgic for the good ol’ days. What days am I talking about? My Data Engineering days when all I had to worry about was reading files with Python and throwing stuff into Postgres or some other database. The good ol’ days. The other day I was reminiscing about what I worked on a lot during the beginning of my data career. Relational databases plus Python was pretty much the name of the game.
One of the struggles I always had was how fast can I load this data into Postgres? psycopg2 was always my Python package of choice for working with Postgres, it’s a wonderful library. Today I want to give a shout-out to my old self by performance testing Python inserts into Postgres. There are about a million ways and sizes and shapes to getting a bunch of records from some CSV file, through Python, and into Postgres.
I also enjoy making people mad … there’s always that. Nothing makes people mad at you like a good ol’ performance test 🙂
Postgres + Python = psycopg2
I’m sure you all have your favorite crappy Python packages for working with databases, but there is only one choice for Postgres … psycopg2. It can’t be beaten.
Let’s get right to it, I don’t want to spend a lot of time on the basics, but I must spend a little. There are a few topics you need to know about when using psycopg2 in Python, and without further ado, here they are.
- connection
- error catching and handling
- cursors
- execute
I wrote a little diddy about Postgres and Python a few moons ago, here is a sample from a simple program.
import psycopg2
host = 'localhost'
database = 'my_database'
user = 'postgres'
pass = 'postgres'
try:
conn = psycopg2.connect(host=host, database=database, user=user, password=pass)
except psycopg2.Error as e:
print(f'Had problem connecting with error {e}.')
database_cursor = conn.cursor()
query = 'CREATE TABLE books (id INT NOT NULL, author varchar(150), title varchar(50));'
try:
database_cursor.execute(query)
conn.commit()
except psycopg2.Error as e:
print(f'Problem executing query {query} with error {e}.')
conn.rollback()The connection gives you access to the database. The cursor which is created from that connection gives you a transactional method to submit queries through that connection to the database. execute is where your logic and SQL go. The try and except are always important in this Python + Postgres world so when something goes wrong, you actually get an idea back from the database about what failed.
Anyways, enough with the boring stuff let’s get to the problem that most people end up dealing with at some point.
The classic Data Engineering problem at the small scale is that you have a bunch of records, typically in a flat file or CSV file and the records need to end up into Postgres somehow or some way.
Python INSERTS into Postgres.
There are a few ways to INSERT records into Postgres with psycopg2, let’s list them. All code is available on GitHub.
- execute()
- executemany()
- execute_values()
- mogrify()
I’m sure some angry person will email me another way.
I set up a few things to do this, mainly a Dockerfile and docker-compose to run Postgres and create a table needed for the testing.
Here is a sample docker-compose file that by running docker-compose up will run a local Postgres instance.
version: '3.7'
services:
postgres:
image: postgres:10.5
restart: always
environment:
- POSTGRES_USER=postgres
- POSTGRES_PASSWORD=postgres
ports:
- '5432:5432'
volumes:
- ./postgres-data:/var/lib/postgresql/data
- ./sql/create_tables.sql:/docker-entrypoint-initdb.d/create_tables.sqlAlso, as you can see, we need a CREATE script for a SQL table. We are going to use the Divvy Bike trip-free dataset, as our records for the insert. Here is a CREATE to hold those records. This file goes into the sql/create_tables.sql location on your machine.
CREATE TABLE trip_data
(
ride_id VARCHAR,
rideable_type VARCHAR,
started_at TIMESTAMP,
ended_at TIMESTAMP,
start_station_name VARCHAR,
start_station_id INT,
end_station_name VARCHAR,
end_station_id INT,
start_lat VARCHAR,
start_lng VARCHAR,
end_lat VARCHAR,
end_lng VARCHAR,
member_casual VARCHAR
);I know, nothing fancy here. I also grabbed the 2021 Divvy bike trip data.
Postgres Python INSERT
Again, here is the list of options we will try with psycopg2 …
- execute()
- executemany()
- execute_values()
- mogrify()
Here is the code I’m going to use to loop the files and insert the records. We are going to try to execute() first.
import psycopg2
import csv
from glob import glob
from datetime import datetime
def connect_postgres(host: str = 'localhost', user: str = 'postgres', pwd: str = 'postgres', port: int = 5432):
try:
conn = psycopg2.connect(f'postgresql://{user}:{pwd}@{host}:{port}')
return conn
except psycopg2.Error as e:
print(f'Had problem connecting with error {e}.')
def pull_data_files(loc: str = 'data/*csv') -> list:
files = glob(loc)
return files
def read_file(file: str) -> list:
with open(file) as csvfile:
reader = csv.reader(csvfile)
next(reader, None)
rows = [row for row in reader]
return rows
def insert_rows(rows: list, conn: object) -> None:
cur = conn.cursor()
for row in rows:
cur.execute("INSERT INTO trip_data VALUES (%s, %s, %s, %s, %s, %s, %s, %s, %s, %s, %s, %s, %s)",
(row[0], row[1], row[2], row[3], row[4], row[5], row[6], row[7], row[8], row[9]
, row[10], row[11], row[12]))
conn.commit()
if __name__ == '__main__':
t1 = datetime.now()
conn = connect_postgres()
files = pull_data_files()
for file in files:
print(f"working on file {file}")
rows = read_file(file)
insert_rows(rows, conn)
print(f"finished with file {file}")
conn.close()
t2 = datetime.now()
x = t2 - t1
print(f"time was {x}")
execute() performance
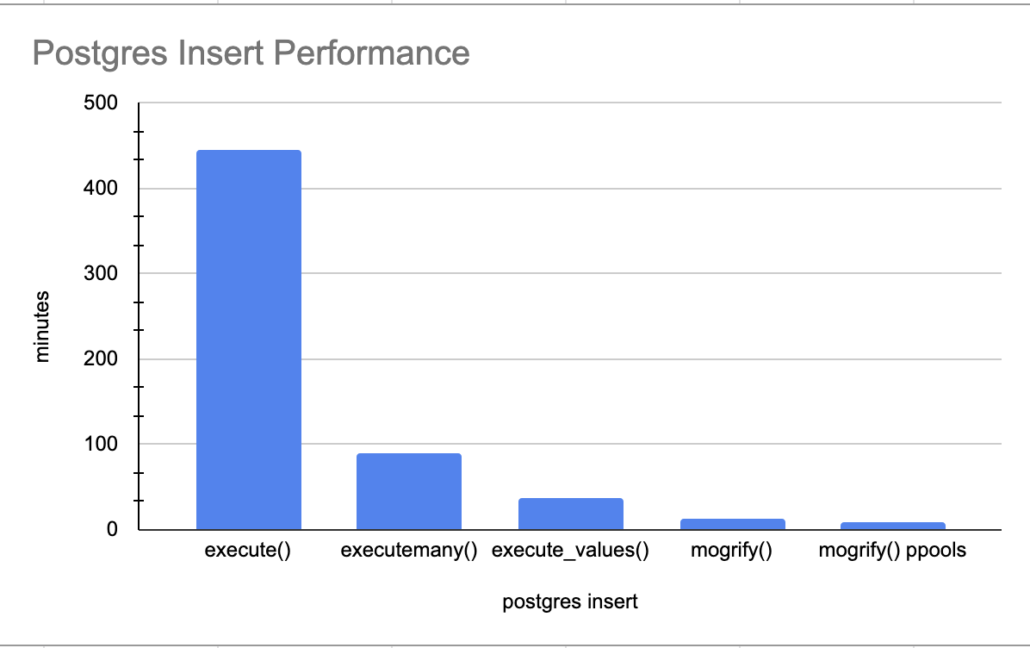
We know this execute() is going to be slow for sure, inserting records one by one. Oh, yes, important note there are 5,347,523 records that get inserted with this dataset.
working on file data/202109-divvy-tripdata.csv
finished with file data/202109-divvy-tripdata.csv
working on file data/202104-divvy-tripdata.csv
finished with file data/202104-divvy-tripdata.csv
working on file data/202107-divvy-tripdata.csv
finished with file data/202107-divvy-tripdata.csv
working on file data/202111-divvy-tripdata.csv
finished with file data/202111-divvy-tripdata.csv
working on file data/202101-divvy-tripdata.csv
finished with file data/202101-divvy-tripdata.csv
working on file data/202102-divvy-tripdata.csv
finished with file data/202102-divvy-tripdata.csv
working on file data/202103-divvy-tripdata.csv
finished with file data/202103-divvy-tripdata.csv
working on file data/202110-divvy-tripdata.csv
finished with file data/202110-divvy-tripdata.csv
working on file data/202106-divvy-tripdata.csv
finished with file data/202106-divvy-tripdata.csv
working on file data/202108-divvy-tripdata.csv
finished with file data/202108-divvy-tripdata.csv
working on file data/202105-divvy-tripdata.csv
finished with file data/202105-divvy-tripdata.csv
time was 7:26:07.847687Yikes. Slowwww….. 7 hours for 5.3 million records. I excepted bad, but not that bad.
executemany() performance
Time to try the old exectuemany() statement. This method accepts an list of lists or an sequence of senquences. So to do this I’m going to add two new methods to my code, one to break my data frame rows up into evenly sized chunks.
Chunker method.
def chunker(lst, n):
chunks = [lst[i * n:(i + 1) * n] for i in range((len(lst) + n - 1) // n )]
return chunksNew logic to insert the chunks with executemany()
for file in files:
print(f"working on file {file}")
rows = read_file(file)
chunks = chunker(rows, 50000)
for chunk in chunks:
insert_rows(chunk, conn)We also have to change our INSERT up slightly.
def insert_rows(rows: list, conn: object) -> None:
cur = conn.cursor()
inputs = [[row[0], row[1], row[2], row[3], row[4], row[5], row[6], row[7], row[8], row[9]
, row[10], row[11], row[12]] for row in rows]
cur.executemany("INSERT INTO trip_data VALUES (%s, %s, %s, %s, %s, %s, %s, %s, %s, %s, %s, %s, %s)", inputs)
conn.commit()Here are the numbers for the executemany()
working on file data/202109-divvy-tripdata.csv
finished with file data/202109-divvy-tripdata.csv
working on file data/202104-divvy-tripdata.csv
finished with file data/202104-divvy-tripdata.csv
working on file data/202107-divvy-tripdata.csv
finished with file data/202107-divvy-tripdata.csv
working on file data/202111-divvy-tripdata.csv
finished with file data/202111-divvy-tripdata.csv
working on file data/202101-divvy-tripdata.csv
finished with file data/202101-divvy-tripdata.csv
working on file data/202102-divvy-tripdata.csv
finished with file data/202102-divvy-tripdata.csv
working on file data/202103-divvy-tripdata.csv
finished with file data/202103-divvy-tripdata.csv
working on file data/202110-divvy-tripdata.csv
finished with file data/202110-divvy-tripdata.csv
working on file data/202106-divvy-tripdata.csv
finished with file data/202106-divvy-tripdata.csv
working on file data/202108-divvy-tripdata.csv
finished with file data/202108-divvy-tripdata.csv
working on file data/202105-divvy-tripdata.csv
finished with file data/202105-divvy-tripdata.csv
time was 1:36:34.174885Well well, 1.5 hours is better than 7, we will take it! On to the next one.
execute_values() performance.
Let’s try the newer execute_values(), it’s pretty easy to change and just swap it out for the execute_many(). I’m honestly not sure what the difference is.
def insert_rows(rows: list, conn: object) -> None:
cur = conn.cursor()
inputs = [[row[0], row[1], row[2], row[3], row[4], row[5], row[6], row[7], row[8], row[9]
, row[10], row[11], row[12]] for row in rows]
cur.execute_values("INSERT INTO trip_data VALUES (%s, %s, %s, %s, %s, %s, %s, %s, %s, %s, %s, %s, %s)", inputs)
conn.commit()Well, look at that.
working on file data/202109-divvy-tripdata.csv
finished with file data/202109-divvy-tripdata.csv
working on file data/202104-divvy-tripdata.csv
finished with file data/202104-divvy-tripdata.csv
working on file data/202107-divvy-tripdata.csv
finished with file data/202107-divvy-tripdata.csv
working on file data/202111-divvy-tripdata.csv
finished with file data/202111-divvy-tripdata.csv
working on file data/202101-divvy-tripdata.csv
finished with file data/202101-divvy-tripdata.csv
working on file data/202102-divvy-tripdata.csv
finished with file data/202102-divvy-tripdata.csv
working on file data/202103-divvy-tripdata.csv
finished with file data/202103-divvy-tripdata.csv
working on file data/202110-divvy-tripdata.csv
finished with file data/202110-divvy-tripdata.csv
working on file data/202106-divvy-tripdata.csv
finished with file data/202106-divvy-tripdata.csv
working on file data/202108-divvy-tripdata.csv
finished with file data/202108-divvy-tripdata.csv
working on file data/202105-divvy-tripdata.csv
finished with file data/202105-divvy-tripdata.csv
time was 0:37:03.791394execute_values() is way faster than that executemany()! 37 minutes for 5.3 million records. I would say that’s more like it!
mogrify() performance.
Last but not least, the strangely named mogrify(). It’s the little weird cousin when it comes to Python Postgres inserts. It basically buildings query strings exactly that they would be when send to the database. It requires you to have a list of tuples, here are my updated functions to handle this.
def read_file(file: str) -> list:
with open(file) as csvfile:
reader = csv.reader(csvfile)
next(reader, None)
rows = [tuple(row) for row in reader]
return rows
def insert_rows(rows: list, conn: object) -> None:
cur = conn.cursor()
args_str = ','.join(cur.mogrify("(%s,%s,%s,%s,%s,%s,%s,%s,%s,%s,%s,%s,%s)", row).decode('utf8') for row in rows)
cur.execute("INSERT INTO trip_data VALUES " + args_str)
conn.commit()Slight changes as you can see with the tuples() inside the list comprehension. Also, you can see the somewhat strange layout of the mogrify method itself.
Well that blew my socks off.
working on file data/202109-divvy-tripdata.csv
finished with file data/202109-divvy-tripdata.csv
working on file data/202104-divvy-tripdata.csv
finished with file data/202104-divvy-tripdata.csv
working on file data/202107-divvy-tripdata.csv
finished with file data/202107-divvy-tripdata.csv
working on file data/202111-divvy-tripdata.csv
finished with file data/202111-divvy-tripdata.csv
working on file data/202101-divvy-tripdata.csv
finished with file data/202101-divvy-tripdata.csv
working on file data/202102-divvy-tripdata.csv
finished with file data/202102-divvy-tripdata.csv
working on file data/202103-divvy-tripdata.csv
finished with file data/202103-divvy-tripdata.csv
working on file data/202110-divvy-tripdata.csv
finished with file data/202110-divvy-tripdata.csv
working on file data/202106-divvy-tripdata.csv
finished with file data/202106-divvy-tripdata.csv
working on file data/202108-divvy-tripdata.csv
finished with file data/202108-divvy-tripdata.csv
working on file data/202105-divvy-tripdata.csv
finished with file data/202105-divvy-tripdata.csv
time was 0:13:43.270913Only a short 13 minutes for the insert of 5.3 million records. Now there is nothing to complain about with that is there!!
But wait there is more, just like late night TV informercials. Can we beat this number by using ProcessPoolExectuor aka multiple cores to insert file records in parallel, just to see how fast we get it it going? How can we accomplish this?
Let’s wrap all the logic of the INSERT into a one big function.
def file_insert(file: str):
conn = connect_postgres()
print(f"working on file {file}")
rows = read_file(file)
chunks = chunker(rows, 50000)
for chunk in chunks:
insert_rows(chunk, conn)
print(f"finished with file {file}")Once that is done, let’s just create a ProcessPool and map all the files too it.
if __name__ == '__main__':
t1 = datetime.now()
files = pull_data_files()
with ProcessPoolExecutor(max_workers=5) as poolparty:
poolparty.map(file_insert, files)
t2 = datetime.now()
x = t2 - t1
print(f"time was {x}")Even better, cut the time in half, a little under 8 minutes.
working on file data/202101-divvy-tripdata.csv
working on file data/202111-divvy-tripdata.csv
working on file data/202104-divvy-tripdata.csv
working on file data/202107-divvy-tripdata.csv
working on file data/202109-divvy-tripdata.csv
finished with file data/202101-divvy-tripdata.csv
working on file data/202102-divvy-tripdata.csv
finished with file data/202102-divvy-tripdata.csv
working on file data/202103-divvy-tripdata.csv
finished with file data/202104-divvy-tripdata.csv
working on file data/202110-divvy-tripdata.csv
finished with file data/202111-divvy-tripdata.csv
working on file data/202106-divvy-tripdata.csv
finished with file data/202103-divvy-tripdata.csv
working on file data/202108-divvy-tripdata.csv
finished with file data/202107-divvy-tripdata.csv
working on file data/202105-divvy-tripdata.csv
finished with file data/202109-divvy-tripdata.csv
finished with file data/202110-divvy-tripdata.csv
finished with file data/202106-divvy-tripdata.csv
finished with file data/202108-divvy-tripdata.csv
finished with file data/202105-divvy-tripdata.csv
time was 0:07:54.780810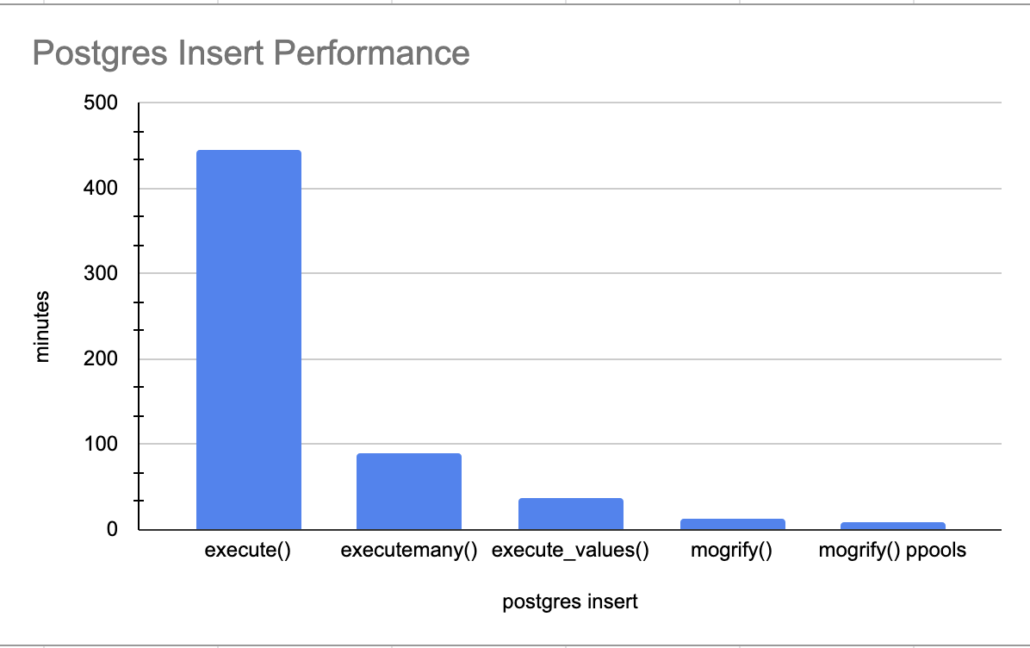
Musings
I guess it pays off to do your homework. It’s been awhile since I’ve played around with Python Postgres inserts with psycopg2. When you play all day long with PySpark, you forget about the good old days with relational databases, and the challenges of storing and moving data with Python in and out of relational databases.
The above graph really illustrates the need to do your research on the Python libraries you use, and get creative. How many people would just wait a few hours for the 5.3 million records to insert and just say “well its a lot of data, what do I do.”
Trail and error is the name of the game, experiment, tweak the code, try new things. Most important of all, combine and mix technologies, be creative. The use of mogrify() and ProcessPoolExecutor() really heated things up, 8 minutes for 5.3 million records isn’t bad. I’m and sure it could be improved upon even more.
Happy hacking.

Also there is the Postgres `COPY` command, which is available in psycopg2 as `cursor.copy_from()` and `cursor.copy_expert()`. You can open your csv and pass the file directly to these methods, they will treat it as if reading from `stdin` (I guess you could make a chain of iterators from your list of files).
Not sure where this would land in your ranking, but pretty sure it beats the `execute` based versions. I had no idea about the benefits of `mogrify` — what does this name even mean? –, so I never even considered it vs. copy. Thanks for bringing it up!)